Network | PHYLOGEOGRAPHIC NETWORK OF SARS-CoV-2 CORONA VIRUS OUTBREAK
Phylogenetic network analysis of SARS-CoV-2 genomes
Forster P, Forster L, Renfrew C, Forster M. Proceedings of the National Academy of Sciences. 8 April 2020
Here you can download the Network files (zip archive) used for the publication.
The publication is based on genome data of 3 March 2020. Download classification of 1001 SARS-CoV-2 virus genome IDs from GISAID, including sampling dates, geographic location of host infection, and their phylogenetic classification into A, B, C types (all full genomes until 24 March 2020). This Excel file lacks sequence information but Dr. Peter Forster can provide another Excel file with mutations with respect to the reference sequence, as follows: According to the GISAID terms, first you need to register with GISAID; then you can contact Dr. Peter Forster and send him proof of being registered at GISAID; then Peter can make the list available to you; please be patient if this takes a few days.
PURPOSES: to help identify "patient zero" (the index patient who was the source of a local outbreak); to help identify relevant virus types for the design of clinical tests, medication and vaccines
DISCLAIMER: This is no medical device or software. For use only at your own risk and your own responsibility. In no event will Fluxus Technology Ltd or its officers be liable for any expenses, losses, or damages resulting from the use or interpretation of the data or the phylogeographic network.
ACKNOWLEDGEMENT: Sequence data were provided by the GISAID COVID-19 sequence database. We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID's EpiFlu(TM) Database on which this research is based. The sequence data contributors are listed in this Excel table.
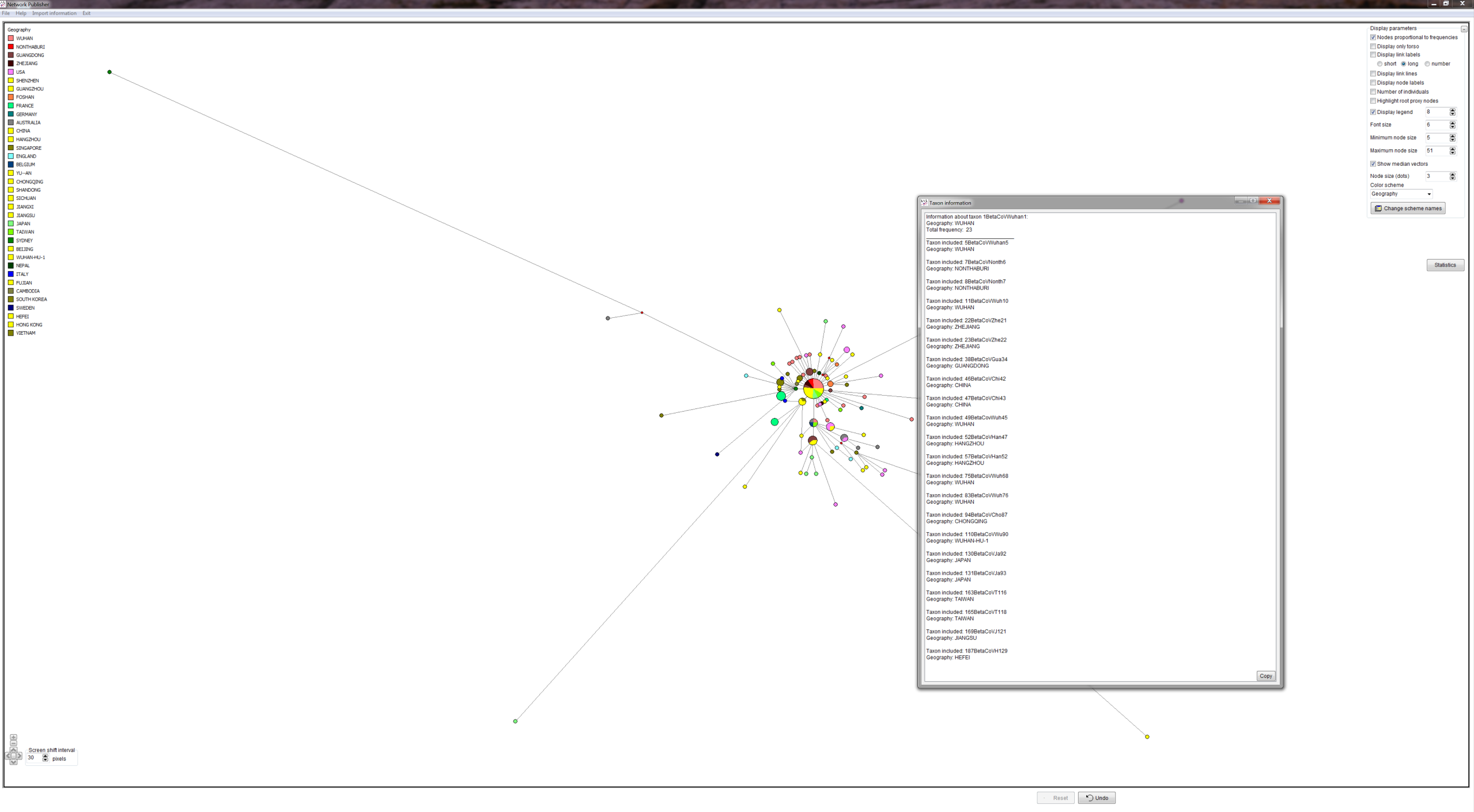
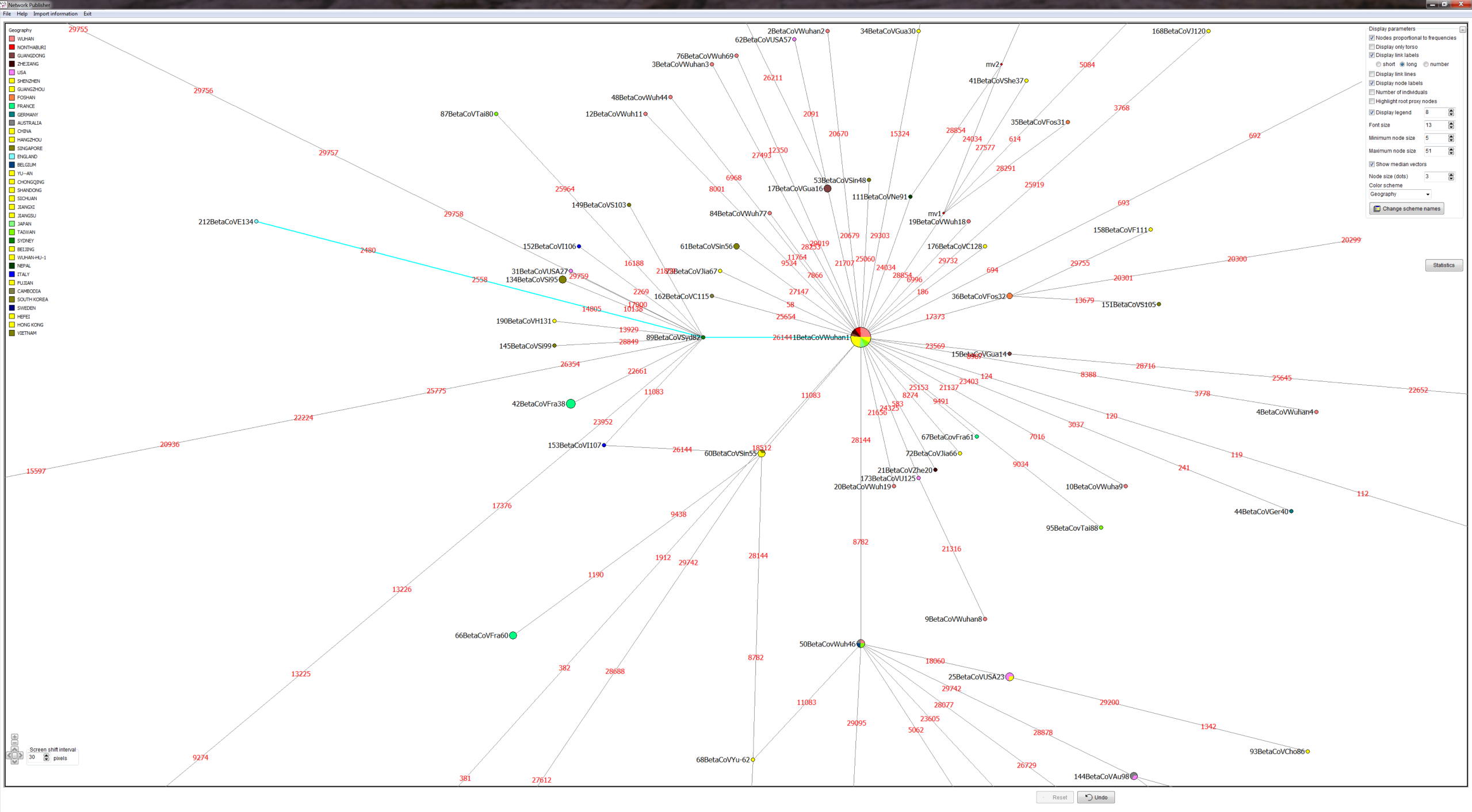
METHODS: GISAID provided COVID-19 virus FASTA sequences. We produced a quality controlled data set, an alignment and phylogenetic network analyses. We produced PDFs and screenshots and Network Publisher fdi files with nodes coloured by geographic origin of virus sequence for interactive exploration. The fdi files can also be read into the free Network Draw program but the geographic colouring is not displayable there.
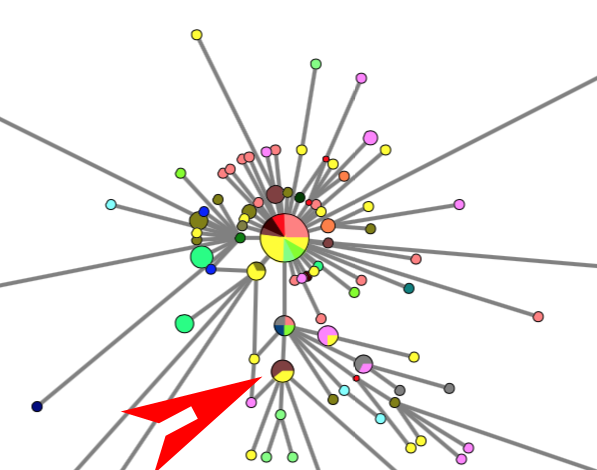
RESULTS: Three main virus types A, B, and C, and their corresponding clusters, can be identified. The root according to the bat corona virus outgroup is in Cluster A, distinguished from the Wuhan-1 reference sequence at positions 8782, 28144, and 29095. The root cluster contains European, Chinese, Japanese and American cases. The analysis and results are described in more detail in the PNAS paper. Download results data (zip file), world phylogeographic network (screenshot), English COVID-19 virus in phylogeographic network (screenshot).
CONTACT: Dr. Peter Forster, pf223 (at) cam (dot) ac (dot) uk
DATE: 29 Feb 2020, UPDATED 8 APRIL 2020